| |
Contents
|
PROGRAM DESCRIPTION
|

|
gff2ps - Produces PostScript graphical output from GFF-files.
gff2ps is a program for visualizing annotations of genomic sequences. The program takes as input the annotated features on a genomic sequence in GFF format, and produces a visual output in PostScript. It can be used in a very simple way, because it assumes that the GFF file itself carries enough formatting information, but it also allows through a number of options and/or a configuration file, for a great degree of customization.
"General Feature Format" (GFF) is described on the Sanger Centre gff definition page.
We will appreciate if you can cite gff2ps paper and/or the URL, as follows:
|
EXAMPLES
|

|
|

|
|
gff2ps has achieved another genome landmark. The mosquito genome annotation for five chromosome arms (2L, 2R, 3L, 3R and X) has been summarized into a two-sided five-pages foldout included as the figure 1 of "The Genome Sequence of the Malaria Mosquito Anopheles gambiae" [Holt et al. Science 298(5591):129-149 (2002)]. Both foldout sides can be downloaded as PDF files from Science web site at the "Annotation of the Anopheles gambiae genome sequence" poster page. You can also download a printable version of this figure from the following links: 1.5Mb gzipped PostScript file or 2.9Mb gzipped PDF file (±196x114cm).
|
|

|
We have performed the map of the Human Genome with gff2ps. 22 autosomic, X and Y chromosomes were displayed in a big poster appearing as the figure 1 of "The Sequence of the Human Genome" [Venter et al. Science 291(5507):1304-1351 (2001)]. The single chromosome pictures can be accessed from here to visualize the web version of the "Annotation of the Celera Human Genome Assembly" poster. A printable version of this figure is also available from the following links: 2.7Mb gzipped PostScript file or 10Mb gzipped PDF file (±107x150cm).
|

|
We have participated on GASP, which results appeared in Genome Research on 24, April. gff2ps generated the poster summarizing all the submitted predictions of each participating group made on the Adh region 2.9Mbp genomic sequence. All those results are analyzed in "Genome Annotation Assesment in Drosophila melanogaster" [Reese et al. Genome Research 10(4):483-501(2000)]. You can also download a printable version of this figure from the following links: 466Kb gzipped PostScript file or 2.2Mb gzipped PDF file (±99x59cm).
|

|
gff2ps was used to obtain the plots for each chromosome arm of Drosophila melanogaster (X, 2L, 2R, 3L, 3R and 4) appearing in the "Coding content of the fly genome" figure, included as a poster in "The Genome Sequence of Drosophila melanogaster" [Adams et al. Science 287(5461):2185-2195(2000)]. A single-page one-side printable version of this figure is also available from the following links: 1.1Kb gzipped PostScript file or 1.7Mb gzipped PDF file (±100x55cm).
|

|
A previous version of gff2ps (0.90) was used to generate the three B0 pages poster for ISMB'99 tutorial #3 on "Drosophila melanogaster ADH region annotation experiment (GASP1)". Each page shows 1 Mb of sequence, splited on four blocks containing all the genomic predictions submitted to that experiment.
You can download here the three panels
poster or
view some of our pictures from ISMB'99 meeting held in Heidelberg in August, 1999.
From this link you can see the last version of the Adh Poster, and you can learn how to get posters with gff2ps.
|
|
|
You can find several examples of the flexibility of gff2ps in those of the following references which include the figure numbers where the program was used.
- gff2ps paper was cited in the following publications:
- M. Stanke et al.
"AUGUSTUS: a web server for gene finding in eukaryotes"
Fig 1, Nucleic Acids Research, 32(Web Server Issue):W309-W312, 2004. [Abstract]
- R.T. Hillman et al.
"An unappreciated role for RNA surveillance"
Fig 4b, Genome Biology, 5(2):R8/1-16, 2004. [Abstract]
- S. Castellano et al.
"Reconsidering the evolution of eukaryotic selenoproteins: a novel non-mammalian family with scattered phylogenetic distribution"
Figs 1A and 2A, EMBO Reports, 5(1):71-77, 2004. [Abstract]
- M. Stanke
"Gene Prediction with a Hidden Markov Model"
Figs 5.2 and 6.2, PhD Thesis.
- Y. Ueno et al.
"Processing sequence annotation data using the Lua programming language."
In M. Gribskov, M. Kanehisa, S. Miyano, and T. Takagi editors:
Genome Informatics 2003. Genome Informatics Series Volume 14. Pp:154-163.
Universal Academy Press, Inc., Tokyo, 2003. [Table of Contents]
- P. Hu and M.R. Brent.
"Using TWINSCAN to predict structures in genomic DNA sequences."
Fig 4.8.5. In A. D. Baxevanis and D. B. Davison, chief editors:
Current Protocols in Bioinformatics. Volume 1 (Supplement 3), Unit 4.8.
John Wiley & Sons Inc., New York, 2002. [Table of Contents]
- J.S. Iacovoni.
"GeneHuggers: database mining and application connectivity tools for subsequence analyses of the human genome."
Bioinformatics, 19(17):2316-2318, 2003. [Abstract]
- G.S. Vernikos et al.
"GeneViTo: Visualizing gene-product functional and structural features in genomic datasets."
BMC Bioinformatics, 4(1):53, 2003. [Abstract]
- R. Gil et al.
"The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes."
Fig 5, PNAS, 100(16):9388-9393, 2003. [Abstract]
- A.K. Hudek et al.
"Genescript: DNA sequence annotation pipeline."
Bioinformatics, 19(9):1177-1178, 2003. [Abstract]
- J.S. Pedersen and J. Hein.
"Gene finding with a hidden Markov model of genome structure and evolution."
Fig 6, Bioinformatics, 19(2):219-227, 2003. [Abstract]
- E. Blanco et al.
"Using geneid to Identify Genes."
Figs 4.3.5, 4.3.7, and 4.3.8. In A. D. Baxevanis and D. B. Davison, chief editors:
Current Protocols in Bioinformatics. Volume 1, Unit 4.3.
John Wiley & Sons Inc., New York, 2002. [Table of Contents]
- N. Rajewsky et al.
"Computational detection of genomic cis-regulatory modules applied to body patterning in the early Drosophila embryo."
Fig 3, BMC Bioinformatics, 3(1):30, 2002. [Abstract]
- R.A. Holt et al.
"The Genome Sequence of the Malaria Mosquito Anopheles gambiae."
Fig 1 (Foldout), Science, 298(5591):129-149, 2002. [Abstract]
- R.J. Mural et al.
"A Comparison of Whole-Genome Shotgun-Derived Mouse Chromosome 16 and the Human Genome."
Fig 2, Science, 296(5573):1661-1671, 2002. [Abstract]
- S. Castellano et al.
"In silico identification of novel selenoproteins in the Drosophila melanogaster genome."
Figs 1A and 2A, EMBO Reports, 2(8):697-702, 2001. [Abstract]
- J.C. Venter et al.
"The Sequence of the Human Genome."
Fig 1 (Companion Poster), Science, 291(5507):1304-1351, 2001. [Abstract]
- R. Guigó et al.
"An Assessment of Gene Prediction Accuracy in Large DNA Sequences."
Figs 2 and 3A+B+C, Genome Research 10(10):1631-1642, 2000. [Abstract]
- M.G. Reese et al.
"Genome Annotation Assessment in Drosophila melanogaster."
Companion Poster, Genome Research, 10(4):483-501, 2000. [Abstract]
- M.D. Adams et al.
"The Genome Sequence of Drosophila melanogaster."
Fig 4 (Foldout), Science, 287(5461):2185-2195, 2000. [Abstract]
- R. Guigó et al.
"Sequence Similarity Based Gene Prediction."
Figs 1, 2, and 3. In S. Suhai editor:
Genomics and Proteomics: Functional and Computational Aspects. Pp:95-105.
Plenum Publishing Corporation, 2000. [Table of Contents]
- gff2ps URL was cited in the following publications:
- M. Alexandersson et al.
"SLAM: cross-species gene finding and alignment with a generalized pair hidden Markov model."
Fig 1, Genome Research, 13(3):496-502, 2003. [Abstract]
- B.P. Berman et al.
"Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome."
Figs 1 and 4A, PNAS, 99(2):757-762, 2002. [Abstract]
- A. Kozik et al.
"GenomePixelizer–a visualization program for comparative genomics within and between species."
Bioinformatics, 18(2):335-336, 2002. [Abstract]
- S. Lewis et al.
"Annotating eukaryote genomes."
Curr Opin Struct Biol, 10(3):349-54, 2000. [Abstract]
- T. Thomson et al.
"Fusion of the human gene for the polyubiquitination co-effector UEV-1 with kua, a newly identified gene."
Figs 2C+E and 3A+B, Genome Research, 10(11):1743-1756, 2000. [Abstract]
- G. Parra et al.
"Geneid in Drosophila."
Fig 1, Genome Research, 10(4):511-515, 2000. [Abstract]
- gff2ps was used, but not cited (sigh!), in the following publications:
- R. Guigó et al.
"Comparison of mouse and human genomes followed by experimental verification yields an estimated 1,019 additional genes."
Fig 2, PNAS, 100(3):1140-1145, 2003. [Abstract]
- Mouse Genome Sequencing Consortium.
"Initial sequencing and comparative analysis of the mouse genome."
Fig 16, Nature, 420(6915):520-562, 2002. [Abstract]
Do you think we have forgotten any citation ? Do not hesitate to send an email with the corresponding citation to authors and we will include it here...
|
HOWTOs
|

|
In this section you can find usefull tutorials on how to use gff2ps. It will be regularly updated with new documents.
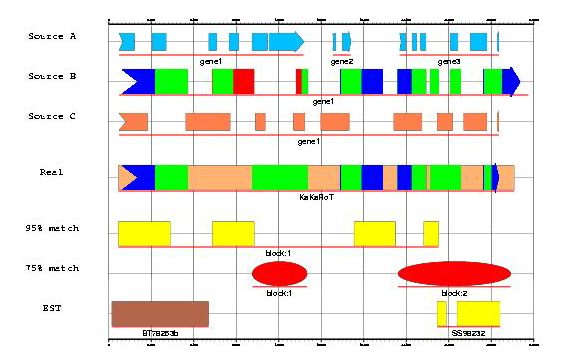
- Comparing sources with gff2ps:
One of the gff2ps program strenghts is comparing results from different sources, so it is easy to see differences among a genomic sequence annotation and one or more gene prediction programs, including results from other programs such blast.
- Visualizing PostScript output from gff2ps:
gff2ps and gff2aplot programs produce PostScript plots, in this howto we will try to provide some help on handling that PostScript output and converting to other formats (including bitmaps and PDF).
- Using gff2ps to visualize gene predictions from geneid:
The following book chapter describes a protocol to run geneid (http://genome.crg.es/software/geneid/) on DNA sequences and describes many of its features. Moreover, there are several command-line examples of how to process the gene prediction results and plot them with gff2ps.
E. Blanco, G. Parra and R. Guigó. "Using geneid to Identify Genes."
In A. D. Baxevanis and D. B. Davison, chief editors: Current Protocols in Bioinformatics. Volume 1, Unit 4.3.
John Wiley & Sons Inc., New York, 2002. ISBN: 0-471-25093-7. [Table of Contents]
You are welcome to provide more examples on how did you use gff2ps in your projects, by sending your report files or a link to your own html report/howto. Your experience will be valuable for other users, mostly for newer ones. Send an email to authors, we will try to include here your contribution as soon as possible.
|
NEWS
|

|
| |
27 May 2003 |
v0.98 |
Minor changes (upgrading to v0.98d):
+ A bug when setting zoom area from command-line has been fixed. Thanks to Massimo Vergassola, who noticed us that issue.
+ Rainbow fill for shapes now depends on feature scores, so color is computed from that GFF field in a continuous color gradient (violet-blue-cyan-green-yellow-orange-red, from lower up to higher scores respectively).
+ PostScript variables defining offsets for axes coordinates have been made global.
|
|
| |
08 Nov 2002 |
v0.98 |
Minor changes (upgrading to v0.98c):
+ Custom file "zoom" option now is not overridden by command-line zoom.
+ Sorting overlapping features for PS layers has been reviewed.
|
|
| |
04 Oct 2002 |
v0.98
|
gff2ps was used on the visualization of the malaria mosquito genome annotation appearing in Science. Up to 278Mbp length, five chromosome arms (2L, 2R, 3L, 3R and X), scaffolds mapped on those chromosomes, a genic set close to 14000 genes, chromosome level polymorphisms, gene expression levels data, homology to Drosophila, SNPs density and G+C content, were integrated into this figure.
|
|
| |
17 Jul 2002 |
v0.98 |
Minor changes (upgrading to v0.98b):
+ Fixing a silly mistake on landscape margins definition in PS code for v0.98, that was making the PS files to crash in that mode.
+ Group color filling gawk function was also checked to be able to set group shape color properly for the PS code.
|
|
| |
04 Jul 2002 |
v0.98 |
v0.98 has been released, it includes few small fixes. Thanks to Gengxin Chen, who pointed that the regular expresion we were using for parsing the strand field from the GFF records will fail on some systems.
Nature is publishing the paper describing the sequence and analysis of the chromosome 2 of Dictyostelium discoideum. gff2ps was used by the Genome Sequencing Center at IMB-Jena to produce the set of figures (each of them showing 100Kb of annotated sequence) for the initial for the chromosome 2 map web page, which were provided as supplementary material.
|
|
| |
25 Sep 2001 |
v0.97 |
Two howto documents has been released for gff2ps. v0.97b is now available in our download section, but newest features are not yet documented.
|
|
| |
16 Feb 2001 |
v0.97
|
gff2ps was used to plot the Human Genome annotation appearing in Science. The total sequence length was of 2.90Gb, distributed among the 24 chromosomes (22 autosomic, X and Y). v0.97 will be available in our download section soon.
|
|
| |
24 Mar 2000 |
v0.94 |
gff2ps was used to plot the Drosophila melanogaster whole genome annotation appearing in Science. The total sequence length was of 120Mb, distributed among the six chromosome arms of the four D. melanogaster chromosomes.
|
|
| |
25 Jan 2000 |
v0.94 |
New variable switch to fit feature drawings in the full width source tracks or to split track width, half for drawings and half for feature labels.
Solved a problem with tickmark scale when defining less nucleotides per page than 1000.
Source labels can now be disabled individually.
|
|
| |
17 Jan 2000 |
v0.94 |
A bug for input filename checking fixed.
Vertical pagination had an error on a function counter which is also checked.
|
|
| |
10 Jan 2000 |
v0.94 |
gff2ps WEB SERVER is now available at Institut Pasteur.
A warning for an old variable not used does not appear.
|
|
| |
05 Jan 2000 |
v0.94 |
Fixes a bug on multiple sequences/sources sorting.
Complete "User's Manual" comes with the program (Appendix figures were improved).
|
|
| |
03 Jan 2000 |
v0.93 |
Official ANNOUNCEMENT of first fully operative version of gff2ps.
A previous version bug on overlapping groups distribution on multiple lines is fixed.
Page layout for vertical pagination is also adjusted.
Standard error reports structure improved.
|
|
| |
23 Dec 1999 |
v0.92 |
Finishing the web-pages for the program (this one, the examples page and the ISMB'99 page).
Main GNUawk script is now included onto Main Shell script.
Program can handle input from standard input. Timing section added to standard error reports.
Some enhancements on PostScript prolog section. This code also ends with empty stack.
|
|
| |
27 Sep 1999 |
v0.91 |
Two main pagination functions: one to fit all the source tracks in one physical page, the other one to fix track size and split output in many vertical pages as needed.
PostScript header and prolog variables now are defined within the Main GNUawk script.
First draft of "User's Manual".
|
|
| |
03 Aug 1999 |
v0.90 |
This gff2ps version was used to produce the ADH poster for the ISMB'99 meeting.
The program is based on two scripts: Shell (gff2ps) and GNUawk (gff2ps.awk).
Defined environment variables to set gff2ps.awk directory, the default custom file, and the path for custom files.
|
|
|
DOWNLOADING
|

|
Download from here the latest versions of the user's manual (v0.96) and of the gff2ps program (v0.98l). You must replace from the script the paths for bash and gawk to the ones defined in your system on the following two lines:
| |
#!/your/bin/dir/bash
GAWK="/your/bin/dir/gawk";
|
|
Due to the intensive usage of associative arrays by our program, we recommend to work with GNU awk version greater than 3.0. Also ensure that you have bourne shell in the "/bin/sh", although we recommend to use bash, version 2 or greater.
You can find in our ftp server a gzipped tarball containing the Scripts for gff2ps version 0.94, a README text file, a PostScript Manual and few example drawings. Download this file only if you are interested on the whole example set, the newest versions of the program and the manual are being updated separately and provided as gziped files.
Once you have downloaded files, you can extract them with the following commands, depending on its file extension:
|
`*.gz' |
|
gunzip *.gz |
| `*.tar.gz' |
|
gunzip -c *.tar.gz | tar xvf -
On Linux you can try with: tar zxvf *.tar.gz |
|
REPORTING BUGS
|

|
If you find any bug or something is not plotted properly, you can send a bug report. To easily find what's wrong, you should attach to that e-mail a tarball containing the custom file you were using when the bug ocurred, an example of your input GFF files, the PostScript file generated and a report file that you can get with the "-V" command-line option (type "gff2ps -h" for further info on that verbose mode switch). We will try to answer as soon as possible.
|
WEB SERVER
|

|
Thanks to Catherine Letondal from Institut Pasteur for providing us of a web server for the gff2ps.
That server was made with PISE, a program developed by Catherine to generate web interfaces for molecular biology programs. We should install a mirror in our server as soon as possible.
|
FEATURE LIST
|

|
The following list shows many features of gff2ps.
- Comprehensive plots for any GFF-feature, attributes are defined separately so you can modify only attributes for same file or share same customization among different data-sets.
- All parameters are set by default within the program, but you also can define a default custom file with all your global settings and a extra custom file for small (or big) changes in one plot.
- User-defined custom files can handle regular expressions, allowing you to set the same variable-attribute for multiple GFF-features.
- Program has been defined as a Unix filter so it can handle data from files, redirections and pipes, writing output to standard-output and warnings to standard error.
- Source order is taken from input files, if you swap file or sources order you can visualize tracks with the new input arrangement.
- gff2ps generates hierarchical plots, where higher level is strand -spliting page blocks in many horizontal regions as strands appear in your files [(+)forward/(-)reverse/(.)no_frame]-, then are drawed source and sequence -plot tracks-, followed by groups and lower level provided by GFF-elements.
- Overlapping groups/elements can be displayed in multiple lines, the minimum number of lines to avoid overlapping among all elements.
- Score controls feature width attribute, when is not defined -"."- in GFF-record maximum value for its source is assumed to visualize it.
- features for which frame is specified are plotted using a two color code schema. The upstream half of the graphical element representing the frame of feature and the downstream half the complement modulus three of its remainder. This is useful to check frame consistency between adjacent features (for instance, predicted exons). Two adjacent features are frame-compatible when the color of the downstream half of the upstream feature matches the color of the upstream half of the downstream feature. This two-color code schema, however, is only meaningful when the frame has been defined relative to the feature, and not relative to the sequence.
- gff2ps is able to manage many physical page formats (from A0 to A10, and more -see available page sizes in its manual-), including user-defined ones. This allows, for instance, the generation of poster size genomic maps, or the use of a continuous-paper supporting plotting device, either in portrait or landscape. It's also possible to obtain multiple horizontal and/or vertical pagination.
|
WISH LIST
|

|
Although we have implemented many features, there are some ideas to be added before releasing version 1.0 of gff2ps.
Here is a short list:
- Drawing functions for vector-data to visualize functions, spikes or bar-charts.
- "Splicing" feature to join elements within a group.
- Composite shapes for promoters, restriction enzyme sites, and so on.
- Vertical marks for any desired element to easy view start-end alignment with others.
- Scale rules for any element (now program shows element start and end positions).
- Custom file variables for position, angle and string width for any label.
- Score cut-off option for visualizing features with scores within the defined range.
- Program option to define custom-file variables in command-line.
- Full re-write in Perl...
We are open to any helpful suggestion for improving our program. Do not hesitate to get in touch with us.
|
AUTHORS
|

|
Josep Francesc ABRIL FERRANDO
Roderic GUIGÓ SERRA
CopyRight © 1999 - 2003
gff2ps is under GNU General Public License.
|
|